Dmg Ligand
Feb 28, 2017 Sarcosine, a glycine transporter type 1 inhibitor and an N-methyl-D-aspartate (NMDA) receptor co-agonist at the glycine binding site, potentiates NMDA receptor function. Structurally similar to sarcosine, N,N-dimethylglycine (DMG) is also N-methyl glycine-derivative amino acid and commonly used as a dietary supplement. The present study compared the effects of sarcosine and DMG on NMDA.
A spectrochemical series is a list of ligands ordered on ligand strength and a list of metal ions based on oxidation number, group and its identity. In crystal field theory, ligands modify the difference in energy between the d orbitals (Δ) called the ligand-field splitting parameter for ligands or the crystal-field splitting parameter, which is mainly reflected in differences in color of similar metal-ligand complexes.
Dmg is a strong field ligand or a weak field ligand - Chemistry - Coordination Compounds. Request PDF on ResearchGate Determination of nickel(II) as the nickel dimethylglyoxime complex using colorimetric solid phase extraction Colorimetric solid phase extraction (C-SPE) is an. In silico docking of a DMG molecule into the ligand-binding site of the OpuAC solute receptor. Subtilis OpuAC protein has been crystalized in complex with its ligands glycine betaine (K d of about 20 μM), DMSA (K d of about 100 μM), and proline betaine (K d of about 270 to 300 μM) (54, 55). Number of mixed ligand complexes involving Ni(JI) have been reported and their structures have been determined by single crystal X-ray crystallographi,7. Reports on mixed ligand complexes of planar Ni(lI) with NiS2N2 chromophore are sparse. The present investigation is an extension of our earlier work on mixed ligand complexes8. Dimethylglyoxime (DMG) with molecular formula CH 3 C(NOH)C(NOH)CH 3 is used for the qualitative analysis of nickel and palladium. Its corresponding conjugate base is the complexing agent not the DMG itself. A pair of DMGH − ligand are joined together with hydrogen bonds to give a.
Spectrochemical series of ligands[edit]
The spectrochemical series was first proposed in 1938 based on the results of absorption spectra of cobalt complexes.[1]
A partial spectrochemical series listing of ligands from small Δ to large Δ is given below. (For a table, see the ligand page.)
Gibbed codes legendary elemental team dmg gaige download. O22−< I− < Br− < S2− < SCN− (S–bonded) < Cl− < N3− < F−< NCO− < OH− < C2O42− < H2O < NCS− (N–bonded) < CH3CN < gly (glycine) < py (pyridine) < NH3 < en (ethylenediamine) < bipy (2,2'-bipyridine) < phen (1,10-phenanthroline) < NO2− < PPh3 < CN− < CO
Ligands arranged on the left end of this spectrochemical series are generally regarded as weaker ligands and cannot cause forcible pairing of electrons within the 3d level, and thus form outer orbital octahedral complexes that are high spin. On the other hand, ligands lying at the right end are stronger ligands and form inner orbital octahedral complexes after forcible pairing of electrons within 3d level and hence are called low spin ligands.
However, keep in mind that 'the spectrochemical series is essentially backwards from what it should be for a reasonable prediction based on the assumptions of crystal field theory.'[2] This deviation from crystal field theory highlights the weakness of crystal field theory's assumption of purely ionic bonds between metal and ligand.
The order of the spectrochemical series can be derived from the understanding that ligands are frequently classified by their donor or acceptor abilities. Some, like NH3, are σ bond donors only, with no orbitals of appropriate symmetry for π bonding interactions. Bonding by these ligands to metals is relatively simple, using only the σ bonds to create relatively weak interactions. Another example of a σ bonding ligand would be ethylenediamine, however ethylenediamine has a stronger effect than ammonia, generating a larger ligand field split, Δ.
Ligands that have occupied p orbitals are potentially π donors. These types of ligands tend to donate these electrons to the metal along with the σ bonding electrons, exhibiting stronger metal-ligand interactions and an effective decrease of Δ. Most halide ligands as well as OH− are primary examples of π donor ligands.
Download mac 10.11. Jan 09, 2018 Download Mac OS X El Capitan 10.11 latest bootable DMG image for Macintosh. Mac OS X El Capitan 10.11 is the twelfth major release of Mac OS X now known as macOS. Mac OS X El Capitan 10.11 Review. MacOS X El Captain is the successor of OS X Yosemite providing numerous powerful features enhancements and better performance than the previous releases. Oct 21, 2015 Download OS X El Capitan 10.11.1 Update. The OS X El Capitan 10.11.1 update improves the stability, compatibility, and security of your Mac, and is recommended for all users. This update: Improves installer reliability when upgrading to OS X El Capitan; Improves compatibility with. Load more results. Apple Footer Apple Support. Aug 21, 2019 Mac OS X El Capitan 10.11.1 DMG Mac. Mac OS El Capitan was released to manufacturing on 20th September 2015, almost three and a half years ago. Its latest version 10.11.6 (15G22010) was released on 9th July 2018, almost 7 months ago. It runs on the platform including x86-64.
When ligands have vacant π* and d orbitals of suitable energy, there is the possibility of pi backbonding, and the ligands may be π acceptors. This addition to the bonding scheme increases Δ. Ligands that do this very effectively include CN−, CO, and many others.[3]
Spectrochemical series of metals[edit]
The metal ions can also be arranged in order of increasing Δ, and this order is largely independent of the identity of the ligand.[4]
Mn2+ < Ni2+ < Co2+ < Fe2+ < V2+ < Fe3+ < Cr3+ < V3+ < Co3+
In general, it is not possible to say whether a given ligand will exert a strong field or a weak field on a given metal ion. However, when we consider the metal ion, the following two useful trends are observed:
- Δ increases with increasing oxidation number, and
- Δ increases down a group.[4]
See also[edit]
References[edit]
- Zumdahl, Steven S. Chemical Principles Fifth Edition. Boston: Houghton Mifflin Company, 2005. Pages 550-551 and 957-964.
- D. F. Shriver and P. W. Atkins Inorganic Chemistry 3rd edition, Oxford University Press, 2001. Pages: 227-236.
- James E. Huheey, Ellen A. Keiter, and Richard L. Keiter Inorganic Chemistry: Principles of Structure and Reactivity 4th edition, HarperCollins College Publishers, 1993. Pages 405-408.
Dmg Ligand Strength
- ^R. Tsuchida (1938). 'Absorption Spectra of Co-ordination Compounds. I.'Bull. Chem. Soc. Jpn. 13 (5). doi:10.1246/bcsj.13.388.
- ^7th page of http://science.marshall.edu/castella/chm448/chap11.pdf
- ^Miessler, Gary; Tarr, Donald (2011). Inorganic Chemistry (4th ed.). Prentice Hall. pp. 395–396. ISBN978-0-13-612866-3.
- ^ abhttp://www.everyscience.com/Chemistry/Inorganic/Crystal_and_Ligand_Field_Theories/b.1013.php
| Names | |
|---|---|
| IUPAC name | |
Other names
| |
| Identifiers | |
| |
| ChEMBL | |
| ChemSpider |
|
| ECHA InfoCard | 100.002.201 |
| EC Number | |
PubChemCID | |
| RTECS number |
|
| UNII | |
| |
| |
| Properties | |
| C4H8N2O2 | |
| Molar mass | 116.120 g·mol−1 |
| Appearance | White/Off White Powder |
| Density | 1.37 g/cm3 |
| Melting point | 240 to 241 °C (464 to 466 °F; 513 to 514 K) |
| Boiling point | decomposes |
| low | |
| Structure | |
| 0 | |
| Hazards | |
| Main hazards | Toxic, Skin/Eye Irritant |
| Safety data sheet | External MSDS |
| GHS pictograms | |
| GHS Signal word | Danger |
| H228, H301 | |
| P210, P240, P241, P264, P270, P280, P301+310, P321, P330, P370+378, P405, P501 | |
| NFPA 704 (fire diamond) | |
| Related compounds | |
| Hydroxylamine salicylaldoxime | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| verify (what is ?) | |
| Infobox references | |
Dimethylglyoxime is a chemical compound described by the formula CH3C(NOH)C(NOH)CH3. Its abbreviation is dmgH2 for neutral form, and dmgH for anionic form, where H stands for hydrogen. This colourless solid is the dioxime derivative of the diketone butane-2,3-dione (also known as diacetyl). DmgH2 is used in the analysis of palladium or nickel. Its coordination complexes are of theoretical interest as models for enzymes and as catalysts. Many related ligands can be prepared from other diketones, e.g. benzil.
Preparation[edit]
Dmg Ligand
Dimethylglyoxime can be prepared from butanone first by reaction with ethyl nitrite to give biacetyl monoxime. The second oxime is installed using sodium hydroxylamine monosulfonate:[1]
Complexes[edit]
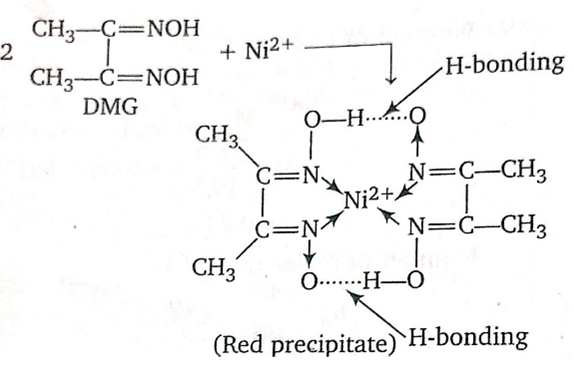
Dimethylglyoxime is used to detect and quantify nickel, which forms the bright red complex nickel bis(dimethylglyoximate) (Ni(dmgH)2). The reaction was discovered by L. A. Chugaev in 1905.[2]
Cobalt complexes have also received much attention. In chloro(pyridine)cobaloxime[3] the macrocycle [dmgH]22− mimics the macrocyclic ligand found in vitamin B12.
References[edit]
- ^Semon, W. L.; Damerell, V. R. (1930). 'Dimethylglyoxime'. Organic Syntheses. 10: 22. doi:10.15227/orgsyn.010.0022.CS1 maint: multiple names: authors list (link)
- ^Lev Tschugaeff (1905). 'Über ein neues, empfindliches Reagens auf Nickel'. Berichte der Deutschen Chemischen Gesellschaft. 38 (3): 2520–2522. doi:10.1002/cber.19050380317.
- ^Girolami, G. S.; Rauchfuss, T.B.; Angelici, R. J. (1999). Synthesis and Technique in Inorganic Chemistry: A Laboratory Manual (3rd ed.). pp. 213–215.